The latest paper by scientists from the Małopolska Centre of Biotechnology of the Jagiellonian University (JU MCB) describes the first high resolution cryo-electron microscopy (cryo-EM) structure of Elongator, which sheds light on the role of the Elp456 subcomplex. The study was published this week in Nucleic Acids Research as a Breakthrough Article.
Scientists from the Max Planck Research Group at the Małopolska Centre of Biotechnology of the Jagiellonian University in Krakow described the role of Elp456, the smaller subcomplex of the Elongator complex, in the tRNA modification pathway. The work of Dr hab. Sebastian Glatt’s team was done in collaboration with the scientists from Kassel University, Berlin Technical University and the European Molecular Biology Laboratory in Hamburg.
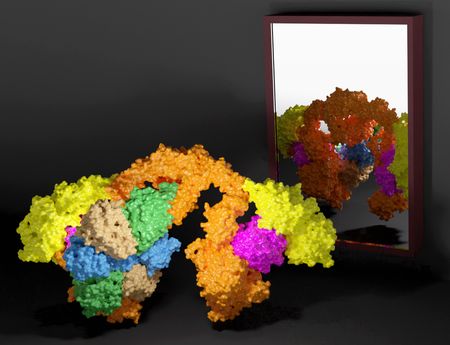
Elongator is a large and multi-subunit (Elp1–6) protein complex with two distinct subcomplexes, namely Elp123 and Elp456. By incorporating carboxymethyl group (cm5), Elongator is responsible for catalyzing the first step of wobble uridine modification at the fifth carbon position which is then further modified by other enzymes. In order to ensure effective co-translational folding of the emerging polypeptide chain, only fully modified tRNAs bind to translating ribosomes in the optimal manner. The team prepared all samples at the MCB Structural Biology Core Facility and collected most of the data at the Titan Krios G3i high-end cryo-electron microscope located at SOLARIS National Synchrotron Radiation Centre. From these data scientists resolved the single particle cryo-EM structures of several intermediates and the fully assembled Elongator complex from yeast and mouse.
The results revealed a very high structural and functional conservation of Elongator among eukaryotes. In vitro and in vivo analyses that were complimentary to the main structural findings allowed for a detailed molecular characterization of the two Elongator subcomplexes and how they interact with each other. “Subtle structural differences between tRNA bound Elp123 and free Elongator allowed us to propose that the function of Elp456 is related to the release of modified tRNAs from the catalytic subcomplex”, adds Dr Marcin Jaciuk, main author of the work. Dr hab. Sebastian Glatt, the last and corresponding author, states that “by comparing the active sites of yeast and mouse Elp123 at different activity states, we managed to take the big next step in characterizing the molecular mechanisms of Elongator’s modification activity. The project that I have started a decade ago during my postdoc times at EMBL has finally reached its destination – understanding how this important protein complex looks and works.”
The paper "Cryo-EM structure of the fully assembled Elongator complex" was supported by the OPUS16 (2018/31/B/NZ1/03559) from the National Science Centre, European Research Council under the European Union’s Horizon 2020 and innovation programme (101001394) of the Foundation for Polish Science (TEAM TECH CORE FACILITY/2017-4/6).